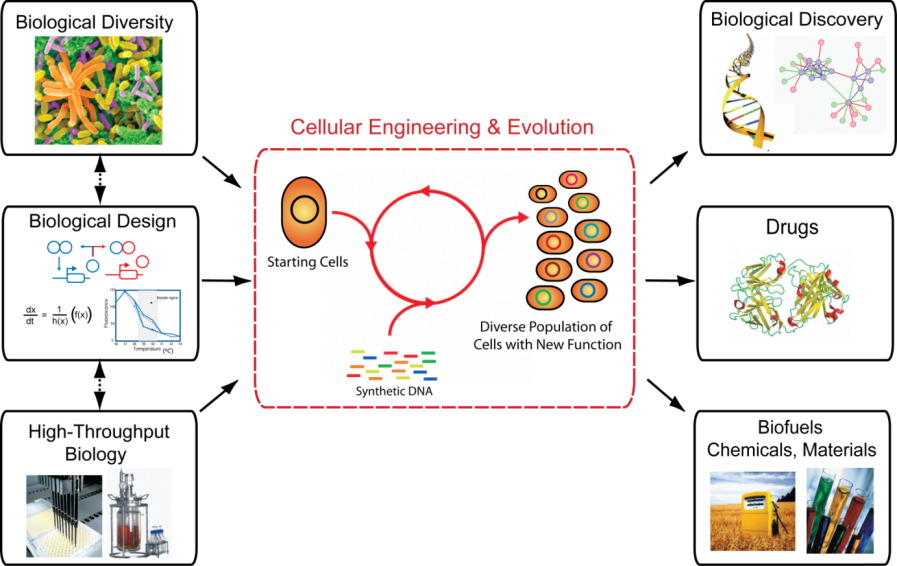
Advances in high-throughput biology and biotechnology have led to an array of novel insights in medicine, agriculture, evolutionary biology and organism diversity. Harnessing biological diversity makes cells ideal for solving global challenges by producing new drugs to treat human disease and generating biologically derived fuels, chemicals and materials to ensure environmental sustainability. Achieving these goals requires a more comprehensive understanding of biological systems that can be leveraged towards the engineering of biosynthetic pathways to generate novel phenotypes. Towards this end, our ability to measure and modify genetic and biochemical components and understand their interactions in pathways, cells and the environment remain defining challenges.
We are focused on developing foundational technologies that enable cellular and genome engineering to understand and modify biological systems. Our approach integrates engineering and evolution through the construction of genes, networks and whole genomes alongside quantitative models to gain a better understanding of whole biological systems. We utilize these insights to uncover new properties of biological systems and generate new phenotypes with the ultimate goal of applying these insights to address global challenges in medicine, energy supply and the environment.
Genome engineering, rapid evolution, new genetic codes
With the advent of next-generation DNA sequencing, our ability to sequence genomes has greatly outpaced our ability to modify genomes. One of the key cellular engineering challenges is the development of high-throughput and automated methodologies for precise manipulation of genomes from single nucleotide to megabase scales. To address these challenges, we have developed methods for versatile genome modification and evolution of cells. Multiplex Automated Genome Engineering (MAGE) simultaneously targets many locations on the chromosome for modification in a single cell or across a population of cells, thus producing combinatorial genomic diversity. Conjugative Assembly Genome Engineering (CAGE) facilitates large-scale hierarchical assembly of genomes with different desired modifications. These methods are broadly being used to program new cellular function, with specific areas of research including the generation of genetic diversity for strain and pathway engineering and the construction of organisms with new genetic codes.
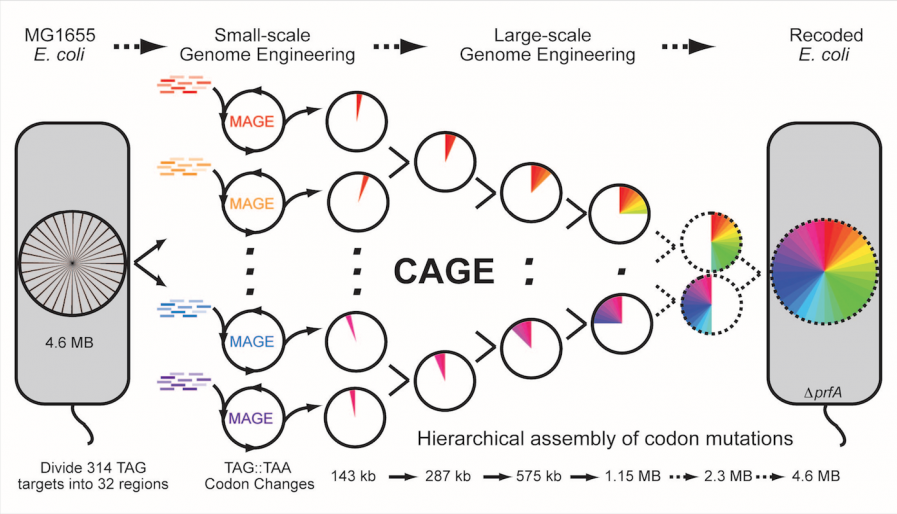
Engineering a new genetic code using MAGE and CAGE
Engineered RNAs
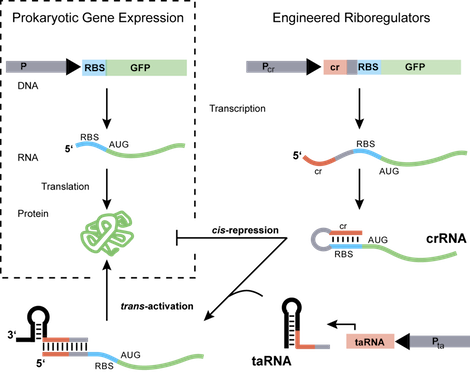
The ability to measure changes in the cellular environment and, in turn, probe and regulate genetic, biochemical or metabolic components remain critical challenges in engineering biological systems. Exploiting the structural and functional diversity of RNA molecules, we design and construct RNA-based systems that can be integrated into biological networks to address these challenges. We build post-transcriptional regulatory systems constructed from highly specific RNA molecules that are functional with any promoter or gene and directly control gene expression in response to a specific biomolecule or chemical input. These engineered riboregulators serve as modular, RNA-based elements in genetic circuits to control and study gene regulation. The design of such functional RNA-based networks could lead to new modes of cellular control and new classes of in vivo probes and sensors that are scalable and applicable to many organisms.
Uncovering complex biological behavior
We integrate systems-specific models to analyze and describe the behavior of engineered genome and RNA research efforts. To complement our engineered genome and RNA efforts, we integrate systems-specific models to analyze and describe the behavior of natural and synthetic networks, genomes and populations. Specifically, we develop systems-specific quantitative models tailored for detailed comparison with the biological systems that we are studying and engineering.